3. Priors with Structural Information#
An example model is defined with a Binomial likelihood and several hierarchical priors:
It is expected that \(p\) should be around 0.5, so the prior on \(p\) is set with \(\alpha = \beta\), since the mean of a Beta distribution is \(k / (k+k) = 0.5\). Lower values of \(k\) result in a wider Beta distribution, while higher values of \(k\) result in a tigher distribution.
The idea is that structural information is contained in the \(p | k \sim \text{Beta} (k,k)\), while priors on \(k\) and \(r\) contain subjective information, which could informative or non-informative.
We can build this model in PyMC:
import pymc as pm
import arviz as az
import matplotlib.pyplot as plt
import numpy as np
with pm.Model() as m:
# priors
r = pm.Beta("r", 2, 2)
k = pm.Geometric("k", r)
p = pm.Beta("p", k, k)
# likelihood
x = pm.Binomial("likelihood", p=p, n=5, observed=3)
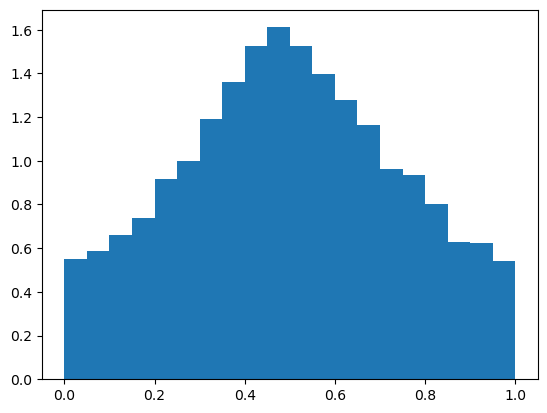
First, let’s sample from the prior predictive and create a histogram of \(p\) before incorporating new data (matching the theoretical plot in the lecture slides):
with m:
trace_prior = pm.sample_prior_predictive(20000)
az.summary(trace_prior)
Sampling: [k, likelihood, p, r]
arviz - WARNING - Shape validation failed: input_shape: (1, 20000), minimum_shape: (chains=2, draws=4)
| mean | sd | hdi_3% | hdi_97% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| r | 0.501 | 0.224 | 0.108 | 0.901 | 0.002 | 0.001 | 19582.0 | 19436.0 | NaN |
| k | 2.946 | 6.119 | 1.000 | 8.000 | 0.044 | 0.031 | 19978.0 | 19825.0 | NaN |
| p | 0.499 | 0.243 | 0.058 | 0.947 | 0.002 | 0.001 | 19934.0 | 19936.0 | NaN |
plt.hist(trace_prior.prior["p"].to_numpy().T, density=True, bins=20)
plt.show()
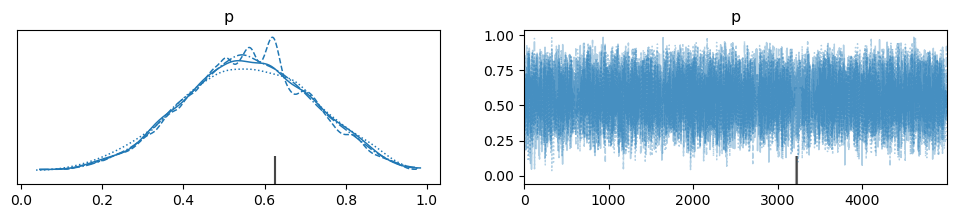
Next, we can sample the posterior distribution. Incorporating the new datapoint of 3/5 successes results in a Bayes estimate in the posterior distribution of \(\approx 0.55\), vs. the MLE of 0.6.
with m:
trace = pm.sample(5000)
az.summary(trace, hdi_prob=0.95)
Multiprocess sampling (4 chains in 4 jobs)
CompoundStep
>NUTS: [r, p]
>Metropolis: [k]
Sampling 4 chains for 1_000 tune and 5_000 draw iterations (4_000 + 20_000 draws total) took 2 seconds.
There were 3 divergences after tuning. Increase `target_accept` or reparameterize.
| mean | sd | hdi_2.5% | hdi_97.5% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| k | 3.090 | 4.111 | 1.000 | 10.000 | 0.227 | 0.170 | 691.0 | 471.0 | 1.01 |
| r | 0.486 | 0.223 | 0.089 | 0.895 | 0.005 | 0.004 | 1559.0 | 938.0 | 1.00 |
| p | 0.557 | 0.158 | 0.260 | 0.873 | 0.001 | 0.001 | 13095.0 | 11443.0 | 1.00 |
az.plot_trace(trace, var_names="p")
plt.show()