import matplotlib.pyplot as plt
import numpy as np
from tqdm.auto import tqdm
7. Metropolis: Normal-Cauchy*#
Adapted from Unit 5: norcaumet.m.
For this example, our model is:
We have a single datapoint, \(x=2\). The professor chose a \(N(x, 1)\) proposal. This is not symmetric around the current state of the chain, so it doesn’t satisfy \(q(\theta^\prime| \theta) = q(\theta | \theta^\prime)\) and we must use Metropolis-Hastings.
Original method#
Here’s how we set up this problem. Assuming we have \(n\) independent data points, the likelihood is:
The Cauchy prior density is:
Then our posterior is proportional to:
When proposing a new value \(\theta'\) from a proposal density \( q(\theta' \mid \theta) \), the acceptance ratio is defined as:
Substituting the expression for the posterior, we get:
Now there are a lot of ways to choose the proposal \(q(\cdot)\). In this case, the professor has chosen one that will simplify our acceptance ratio expression quite a bit, \(N(x, 1)\). I’ll also show how a random walk proposal would work later.
For this example, remember \(n=1\) and \(x=2\).
Since the professor chooses an “independence” proposal, the proposal density is independent of the current state:
The ratio of the proposal densities becomes
Substitute this back into the acceptance ratio:
Use the following fact:
To see that the exponential terms cancel:
This leaves us with a simplified acceptance ratio:
That’s all we need to code our sampler.
Warning
I usually pre-generate arrays of random numbers (see theta_prop and unif variables in the below cell) because it’s computationally faster. However, that only works when those numbers don’t depend on the previous step in the sampling loop.
rng = np.random.default_rng(1)
n = 1000000 # observations
burn = 500
theta = 1 # init
thetas = np.zeros(n)
x = 2 # observed
accepted = 0
# generating necessary randoms as arrays is faster
theta_prop = rng.standard_normal(n) + x
unif = rng.uniform(size=n)
for i in tqdm(range(n)):
r = (1 + theta**2) / (1 + theta_prop[i] ** 2)
rho = min(r, 1)
if unif[i] < rho:
theta = theta_prop[i]
accepted += 1
thetas[i] = theta
thetas = thetas[burn:]
print(f"{np.mean(thetas)=:.3f}")
print(f"{np.var(thetas)=:.3f}")
print(f"{accepted/n=:.3f}")
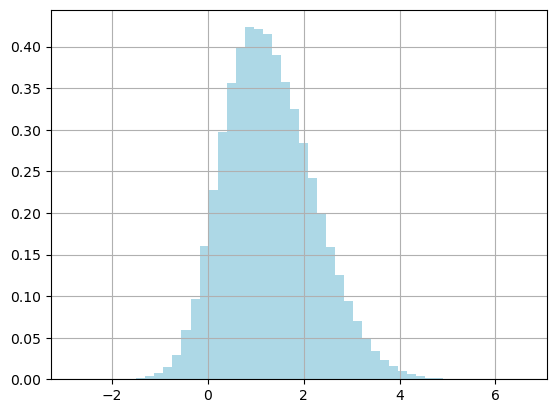
fig, ax = plt.subplots()
ax.grid(True)
plt.hist(thetas, color="lightblue", density=True, bins=50)
plt.show()
np.mean(thetas)=1.283
np.var(thetas)=0.863
accepted/n=0.588
Random-walk proposal#
How would we do this with classic Metropolis? We would use a random-walk proposal, centering the proposal at the previous state of the chain at each step.
Notice I can still pregenerate proposal values, because it’s easy to adjust the pregenerated \(N(0, 1)\) values by adding theta, which centers the distribution at the previous state of the chain.
n = 1000000
burn = 500
theta = 1
thetas = np.zeros(n)
x = 2
accepted = 0
theta_prop = rng.standard_normal(n)
unif = rng.uniform(size=n)
def f(x, theta):
return np.exp(-0.5 * (x - theta) ** 2) / (1 + theta**2)
for i in tqdm(range(n)):
theta_prop_current = theta_prop[i] + theta
r = f(x, theta_prop_current) / f(x, theta)
rho = min(r, 1)
if unif[i] < rho:
theta = theta_prop_current
accepted += 1
thetas[i] = theta
thetas = thetas[burn:]
print(f"{np.mean(thetas)=:.3f}")
print(f"{np.var(thetas)=:.3f}")
print(f"{accepted/n=:.3f}")
fig, ax = plt.subplots()
ax.grid(True)
plt.hist(thetas, color="lightblue", density=True, bins=50)
plt.show()
np.mean(thetas)=1.283
np.var(thetas)=0.865
accepted/n=0.686
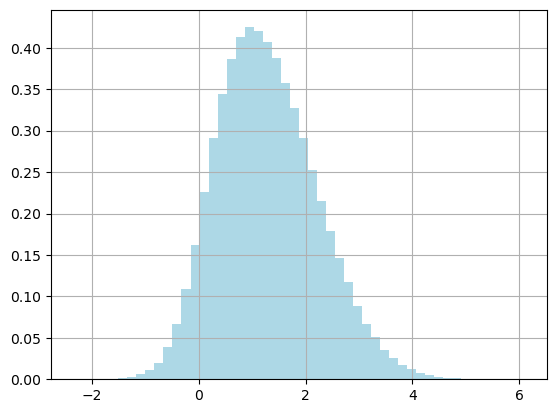
The end results are the same, with a higher acceptance rate! Something to consider: do we always want this behavior from the sampler?
%load_ext watermark
%watermark -n -u -v -iv
Last updated: Thu Feb 13 2025
Python implementation: CPython
Python version : 3.12.7
IPython version : 8.29.0
matplotlib: 3.9.2
tqdm : 4.67.0
numpy : 1.26.4

