import matplotlib.pyplot as plt
import seaborn as sns
import numpy as np
from scipy.stats import gamma, norm
from scipy.optimize import minimize_scalar
sns.set(style="whitegrid")
colors = sns.color_palette("Paired")
4. Laplace’s Method Demo*#
Steps#
As we learned in the last lecture, these are the steps to Laplace approximation:
Find the mode.
The mean of the approximating normal distribution will be the mode, or maximum a posteriori probability (MAP) solution, of the posterior.
We solve for the MAP by maximizing the unnormalized posterior with respect to \(\theta\) or by maximizing the log of the unnormalized posterior:
Find the variance.
In the lecture notation:
Normal approximation:
The Laplace approximation to the posterior is a normal distribution with mean \(\hat{\theta}\) and variance \(Q^{-1}\):
Univariate example#
From Unit 5: laplace.m.
Here’s our example model:
Since this is a conjugate model, we already know the exact posterior is \(Ga(r+\alpha, \beta +x)\). We know the mode is \(\frac{r + \alpha - 1}{\beta + x}\), but let’s pretend we don’t.
Mode#
In this case it’s not too tough to find the mode analytically.
The log of the unnormalized posterior is \(\log g(\theta) = (r + \alpha - 1)\log(\theta) - (\beta + x)\theta\), so we can just take the derivative and set it equal to zero, then solve for \(\theta\):
For this example our given parameters are \(r=20, \alpha=5, \beta=1, x=2\), so our mode is equal to \(8\).
We could also use optimization:
# define our posterior
def neg_log_post(θ, r, α, β, x):
return -((r + α - 1) * np.log(θ) - (β + x) * θ)
r, α, β, x = 20, 5, 1, 2
result = minimize_scalar(
neg_log_post, bounds=(0, 1e6), args=(r, α, β, x), method="bounded"
)
mode = result.x
print(f"Mode: {mode}")
Mode: 8.000001689177818
Variance#
The true variance of the posterior is \(\frac{r+\alpha - 1}{(\beta + x)^2}\).
The approximation distribution’s variance will be:
Compare the true posterior with the approximation#
xx = np.linspace(0, 16, 100000)
variance = (mode**2) / (α + r - 1)
# normal parameters
mu = mode
sigma = variance**0.5
# gamma parameters
shape = r + α
scale = 1 / (β + x)
exact = gamma.pdf(xx, a=shape, scale=scale)
approx = norm.pdf(xx, mu, sigma)
# credible intervals
exact_ci = gamma.ppf(0.025, a=shape, scale=scale), gamma.ppf(
0.975, a=shape, scale=scale
)
approx_ci = norm.ppf(0.025, mu, sigma), norm.ppf(0.975, mu, sigma)
print(f"Exact 95% credible interval: {exact_ci}")
print(f"Laplace approximation 95% credible interval: {approx_ci}")
Exact 95% credible interval: (5.392893949276442, 11.903365864584403)
Laplace approximation 95% credible interval: (4.799393229141485, 11.20061014921415)
Show code cell source
# plotting code
plt.figure(figsize=(10, 6))
plt.plot(xx, exact, label="Exact", color=colors[0])
plt.plot(xx, approx, label="Laplace approx.", color=colors[1])
plt.axvline(x=exact_ci[0], color=colors[2], linestyle="--", label="Exact CI")
plt.axvline(x=exact_ci[1], color=colors[2], linestyle="--")
plt.axvline(x=approx_ci[0], color=colors[3], linestyle="--", label="Approx. CI")
plt.axvline(x=approx_ci[1], color=colors[3], linestyle="--")
plt.axvline(x=mode, color=colors[4], linestyle="-.", label="Mode")
plt.title("Posterior with Laplace Approximation")
plt.xlabel("Value")
plt.ylabel("Density")
plt.ylim(bottom=0)
plt.legend()
plt.show()
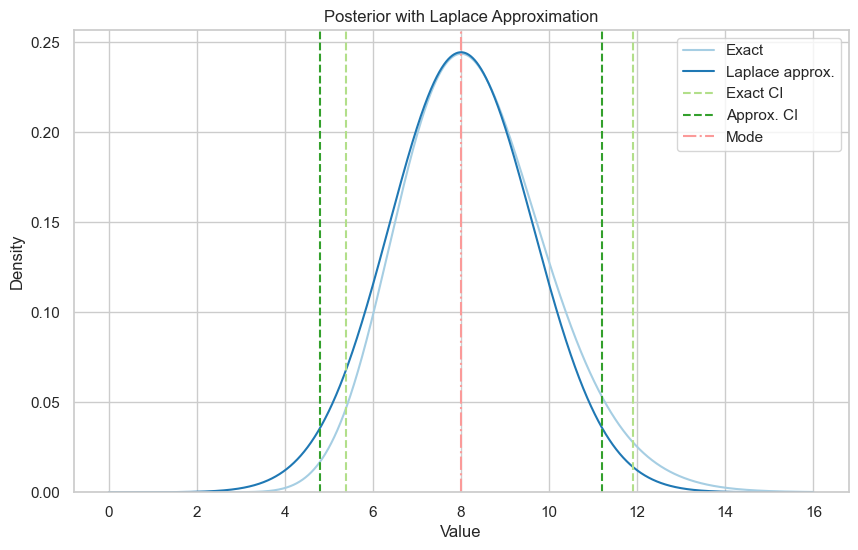
The exact posterior is a bit skewed to the right, so the normal approximation is a bit off, but it’s not too bad.
%load_ext watermark
%watermark -n -u -v -iv
Last updated: Sat Feb 24 2024
Python implementation: CPython
Python version : 3.11.5
IPython version : 8.15.0
numpy : 1.25.2
matplotlib: 3.7.2
seaborn : 0.13.0

